PaVarDB
Genomic Variant Database



PaVarDB is a comprehensive database of genomic variants in Pseudomonas aeruginosa, providing valuable information for clinical studies.
➣ Shows variant data along with resistance phenotype and country information.
➣ Provides visualizations of gene counts and amino acid changes.
➣ Allows users to filter data interactively based on country, antibiotics, strains, genes, and phenotype.
➣ Users can download CSV reports and charts for offline use.
PaVarDB utilizes the Nextflow pipeline, which incorporates bioinformatics tools like Fastp, Snippy, SnpEff, VCFtools, BCFtools, Samtools, and custom bash scripts. The pipeline uses Python to generate an SQLite database. It is adaptable for any bacterial species and can produce annotated VCF and CSV files. To use the pipeline, visit our GitHub repository and follow the manual instructions to download and customize the tools for your specific bacterial species.
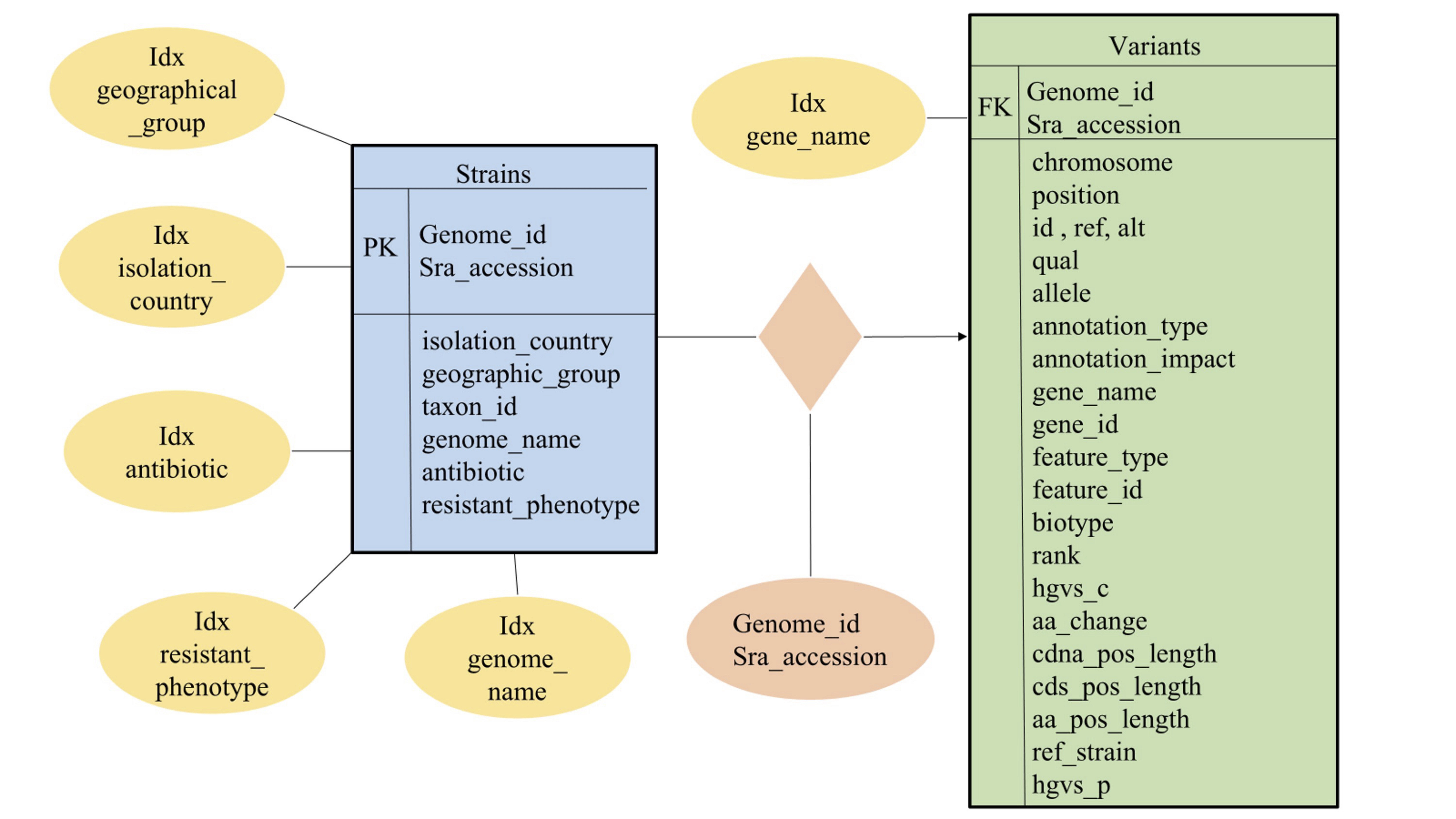
Visit our GitHub repository hereThe PaVarDB database schema is designed to efficiently store and manage genomic variant data in the context of antimicrobial resistance (AMR). It consists of two relational tables: Strains and Variants, which are connected through a composite key consisting of Genome_id and Sra_accession. The Strains table has metadata associated with each bacterial genome, including its geographic origin (isolation_country, geographic_group), taxonomic identifier (taxon_id), genome-specific information (genome_name), the antibiotic (antibiotic), and its observed phenotype (resistant_phenotype). This table uses several indexes to enhance query performance, specifically on fields like geographical_group, isolation_country, antibiotic, resistant_phenotype, and genome_name.
The Variants table stores detailed information about each genomic variant detected in the strains. Each record in this table is linked to the corresponding strain via the shared Genome_id and Sra_accession. The table includes variant-specific attributes such as the chromosome location (chromosome, position), reference and alternate alleles (ref, alt), quality scores (qual), and alleles. It also has annotation information including the gene name and ID, feature type and ID, biotype, variant effect (annotation_type, annotation_impact), and various sequence-level annotations like HGVS notations (hgvs_c, hgvs_p) and amino acid changes (aa_change). The schema supports detailed analysis by also including position and length data for cDNA, CDS, and amino acid changes, as well as reference strain information and also this schema enables a comprehensive representation of strain-level genomic variations and facilitates complex queries linking phenotype, geography, gene function, and variant effects—critical for AMR research. The relational design and indexed fields ensure high performance and scalability for large-scale multi-strain datasets.
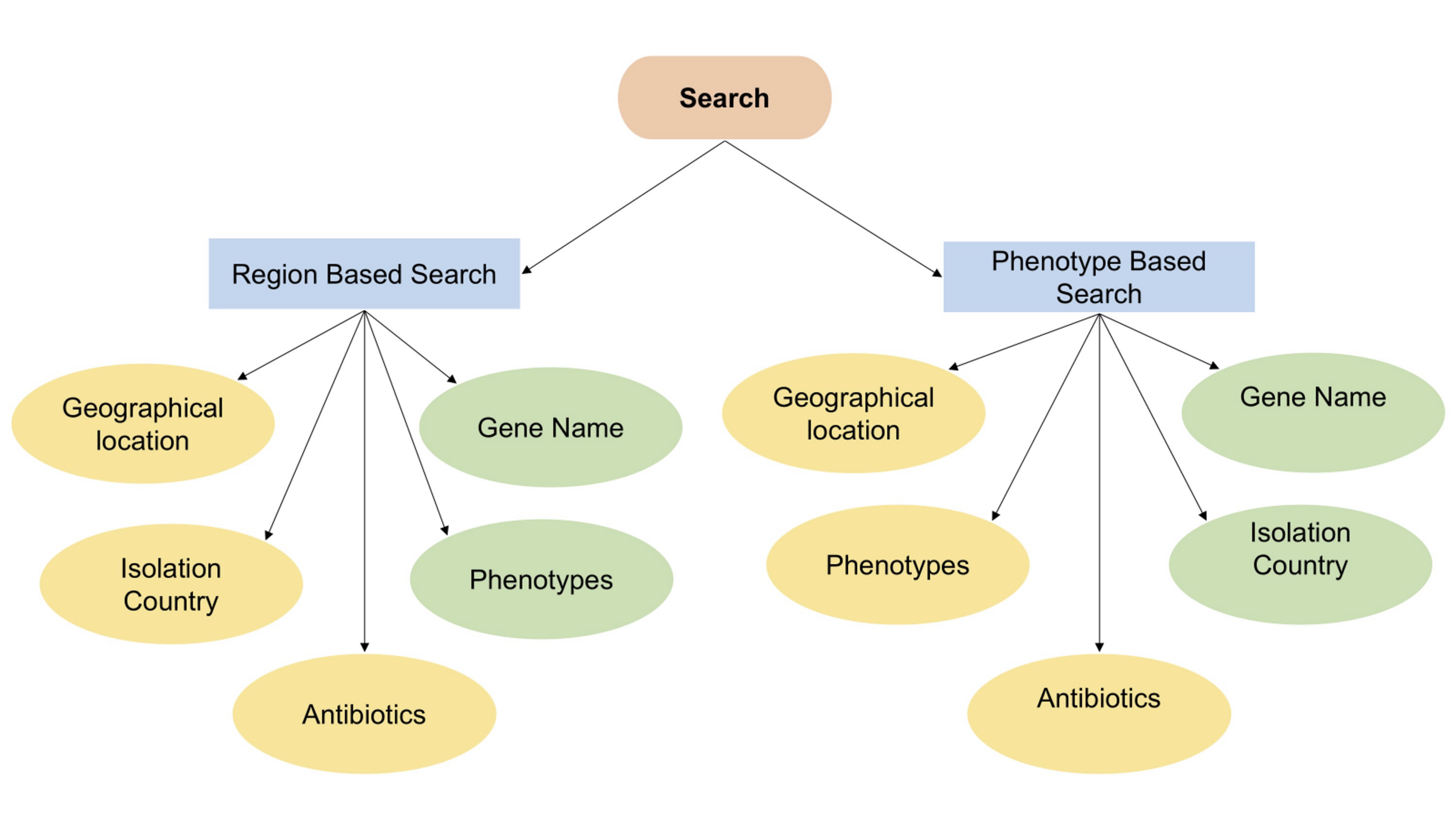
The PaVarDB search system has two main ways to search: Region-Based Search and Phenotype-Based Search. These help users find important information related to clinical studies and antimicrobial resistance more easily. In both methods, some search fields are mandatory (shown in yellow), and some are optional (shown in green).
In Region-Based Search, users must enter the Geographical Location, Isolation Country, and Antibiotic. These help narrow down the search to a specific region and antibiotic resistance. Users can also choose to add Gene Name or Phenotypes to get more specific results. In Phenotype-Based Search, the focus is on whether the bacteria are resistant or susceptible to antibiotics. The required fields are Geographical Location, Phenotypes, and Antibiotic and also users can include Gene Name and Isolation Country to refine the search further.
This simple and flexible search system allows users to explore data from either a regional view or based on resistance, making it useful for researchers and clinical studies.